|
|
BMe Research Grant |
|
George A. Olah Doctoral School of Chemistry and Chemical Technology
Department of Applied Biotechnology and Food Science, BUTE /Institute of Enzymology, RCNS,
Supervisor: Dr. Vértessy G. Beáta
Quantification of Uracil within DNA Using a Sensitive Labeling Method
Introducing the research area
Genome maintenance is a hot topic. Nobel Prize in Chemistry in 2015 was awarded for mechanistic studies of DNA repair. Chromosomes of cells suffer endogenous and exogenous attacks every day, such as UV irradiation, free radicals and carcinogenic agents. DNA lesions can occur also during cell divisions, and due to metabolic processes inside the cells. Hence, DNA repair pathways have exceeding significance in all living creatures.
Uracil is one of the most frequent DNA lesion. Hydrolytic cytosine deamination, resulting in uracil, occurs 100–500 times in a mammalian genome/day [1], and if it is not repaired, causes point mutation. Point mutations can cause cancer and have a role in aging.
Brief introduction of the research place
Beáta G. Vértessy’s Research Group is partly at BUTE and RCNS, HAS: Biostruct Laboratory (BUTE) and Laboratory of Genome Metabolism and Repair (RCNS, HAS) The research activity of Vértessy's Research Group is mainly in vitro and in vivo examination of protein crystallization, structure determination, protein-protein and protein-DNA interaction in mammalian cells and bacteria.
History and context of the research
One of the most frequent lesions in DNA is deoxyuridine. Uracil is one of the four bases in RNA, but the DNA contains thymidine instead of the chemically less stable uracil (Figure 1).

Figure 1: Nucleobases in RNA and DNA
Uracil can incorporate into DNA via two mechanisms: instead of thymine through DNA polymerases or via cytosine deamination. One of the pathways of DNA repair [S1] is base-excision repair (BER). BER and regulation of nucleotide pools are responsible for the prevention of uracil accumulation in DNA. The two key enzymes in this process are uracil-DNA glycosylase (UDG) and dUTPase [S2,S3]. However, uracil also appears in DNA under normal physiological conditions, such as in some heavily uracilated bacteriophages [2,3], and in the genome of HIV [4]. Uracil in DNA is normal intermediate in acquired immunity in human B lymphocytes [5], and emerges in elevated levels in fruitfly larvae, pupae and imago [6]. Chemotherapeutic approaches in cancer treatment also heavily rely on genomic uracil occurrence.
There are some genomic uracil quantification methods in the literature with varying specificity, sensitivity and price [7, 8, 9], see Table 1.
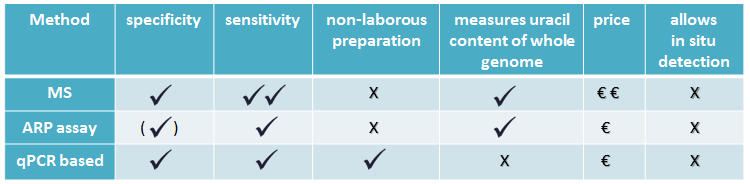
Table 1: Comparing uracil quantification methods
MS-based method is sensitive, but requires quite expensive instrument, and specialized knowledge. Aldehyde reactive probe (ARP) assay measures abasic sites done by UNG, a member of UDG family. A quantitative qPCR based method is suitable for relative quantification.
The research goal, open questions
In the present work we aimed at designing a reliable, fast, cheap and easy method to gain quantitative and qualitative information on uracil levels in DNA.
We developed a uracil sensor, by applying a catalytically inactive mutant of UDG superfamily member UNG, which is capable of binding to, but not excising uracil [S4]. Our sensor has also the potential of using in in situ immunocytochemical approach.
My goal in the first place was to prove that our uracil sensor specifically recognizes uracil, but does not cleave the N-glycosidic bond. Than I had to validate the method on a proper standard and on unknown amount of uracil containing genomic DNA samples.
Methods
For the new uracil quantification method, catalytically inactive UNG was used. Our uracil binding UNG sensor was designed in a way that it can be detected with conventional antibodies in dot-blot applications along with in situ detection using an immunocytochemical approach. The scheme of our method is shown on Figure 2. First of all, DNA is immobilized on nitrocellulose membrane, and labeled with catalytically inactive Flag tagged UNG, then primary (anti Flag) and secondary (horseradish peroxidase coupled anti mouse) antibodies are connected to the complex. Signal capturing is achieved by enhanced chemiluminescence.
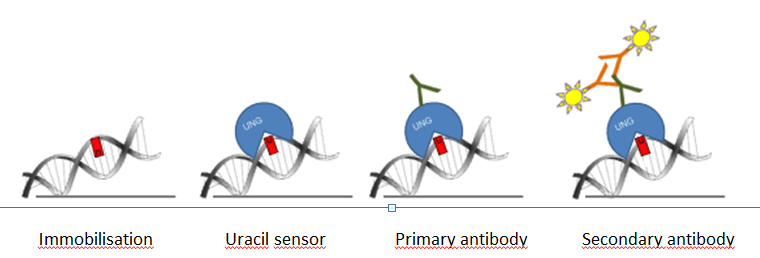
Figure 2: Scheme of the genomic uracil quantification method (dot blot based assay)
We used UNG activity assay to show that our double mutant UNG (D154N, H277N) constructs (due to the point mutations) don’t have uracil excising catalytic activity on uracil-rich DNA. Highly uracilic or “normal” plasmid DNA was treated with wild type (wt) or with mutant UNG constructs, followed by AP endonuclease treatment, that cleaves DNA backbone at base free sites. Samples were analyzed by agarose gel electrophoresis.
Uracil binding capability of the 3xFLAG-ΔUNG construct, used for dot blot, was addressed with electrophoretic mobility shift assay. If UNG molecules bind to uracil containing DNA, the resulting complex will have higher molecular weight, thus appearing on agarose gel at a higher position (gel shift).
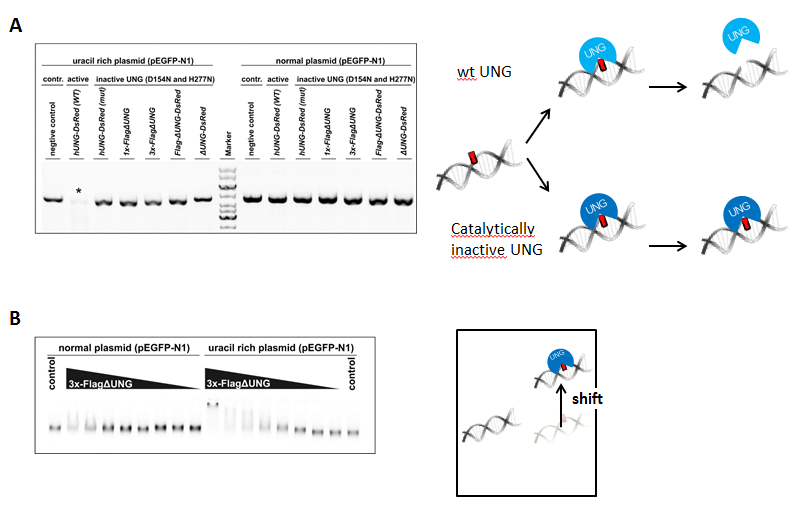
Figure 3: Activity (A) and uracil binding capability (B) of the used UNG constructs
Results
UNG activity assay and electrophoretic mobility shift assay.
On Figure 3 A it is shown that only wild type UNG construct, not harboring the two point mutations, is active on uracil-rich plasmid (signed with asterisk). 3x-FlagΔUNG prefers uracil rich to “normal” DNA. This indicates specific uracil binding affinity (Figure 3 B).
Calibration curve for dot blot based measurements
The catalytically inactive 3xFlag-UNG was used to the dot blot assay. Genomic DNA isolated from log phase growing CJ236 [dut−, ung−] Escherichia coli strain was used as a standard with well-defined, high uracil-content during quantification: Figure 4.
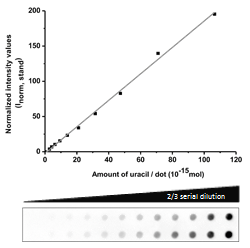
Figure 4: Design of a standard curve for quantification of genomic uracil levels
Measuring genomic uracil content of various biological samples
For each measurement a calibration curve was used, for calculating the uracil levels.
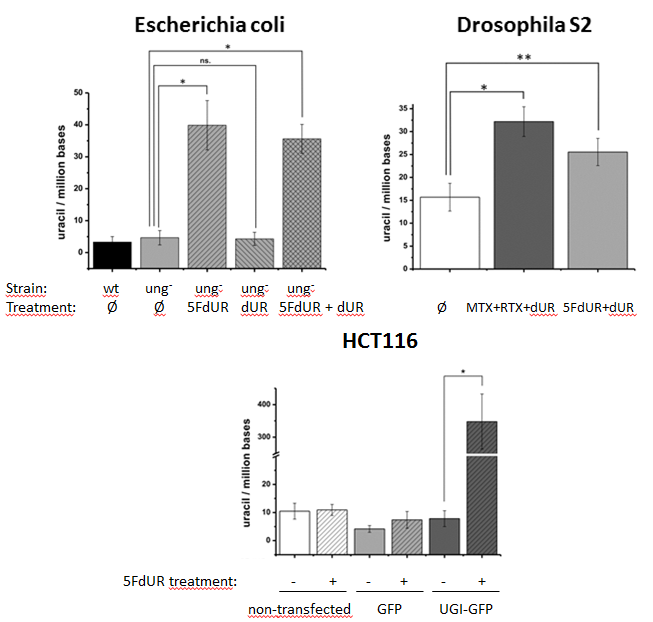
Figure 5: Measurement of genomic uracil content of various species, with or without chemotherapeutic drug treatments (asterisks show significant difference)[S4]
Our uracil sensor was used for quantification of genomic uracil levels of bacteria Escherichia coli, fruit fly cell line S2, and human HCT116 cell line. These cells were also treated with thymidylate synthesis inhibiting chemotherapeutic drugs: 5-fluorodeoxyuridine (5FdUR), methotrexate (MTX), raltitrexed (RTX), and the change in genomic uracil levels were measured. Thymidylate synthase is the enzyme that converts deoxyuridine monophosphate (dUMP) to deoxythymidine monophosphate (dTMP). This is the only de novo dTMP synthesis pathway in all the organisms. In the presence of UNG, elevation of uracil content in DNA could not been detected. However, in the absence of UNG – like an ung- E. coli cell line, the no ung gene coding fruit fly, and in the presence of UNG-inhibitor UGI – the chemotherapeutic drugs increased the uracil content: Figure 5. Adding deoxyuridine (dUR) to the media showed no effect.
Expected impact and further research
My publications in this topic are published in prestigious papers, especially the publication in Nucleic Acids Research, makes up most part of this work, with an impact factor of 9.202 [S4]. Our article on the new uracil quantification method has received considerable international scientific interest: three foreign research groups have got ahold of us (from Norway, India, United Kingdom). In the near future, I would like to measure the genomic uracil level of different developmental stages (e.g. eggs, larvae) of holometabola insects, that all lack ung gene, what is more, in some developmental stages even dUTPase expression is downregulated. I would also like to examine the physiological consequences.
Publications, references, links
Publications (IF: impact factor, IC: independent citations)
[S1] Szalkai B and Scheer I, Nagy K, Vértessy BG, Grolmusz V. The Metagenomic Telescope. PLoS One. 2014 Jul 23;9(7):e101605. (IF: 3.534, IC: 1)
[S2] Róna G, Marfori M, Borsos M, Scheer I, Takács E, Tóth J, Babos F, Magyar A, Erdei A, Bozóky Z, Buday L, Kobe B, Vértessy BG. Phosphorylation adjacent to the nuclear localization signal of human dUTPase abolishes nuclear import: structural and mechanistic insights. Acta Crystallogr D Biol Crystallogr. 2013 Dec;69(Pt 12):2495–505. (IF: 7.232, IC: 8)
[S3] Róna G, Pálinkás HL, Borsos M, Horváth A, Scheer I, Benedek A, Nagy GN, Zagyva I, Vértessy BG. NLS copy-number variation governs efficiency of nuclear import-case study on dUTPases. FEBS J. 2014 Dec;281(24):5463–78. (IF: 3.986, IC: 0)
[S4] Róna G, Scheer I, Nagy K, Pálinkás HL, Tihanyi G, Borsos M, Békési A, Vértessy BG. Detection of uracil within DNA using a sensitive labeling method for in vitro and cellular applications. Nucleic Acids Res. 2016 Feb 18;44(3):e28. ˙(IF: 9.202, IC: 0)
Links.
References.
[1] Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993 Apr 22;362(6422):709–15.
[2] Lozeron,H.A. and Szybalski,W. Incorporation of 5-fluorodeoxyuridine into the DNA of Bacillus subtilis phage PBS2 and its radiobiological consequences. J. Mol. Biol. 1967 Dec 14;30(2):277–90.
[3] Kiljunen S., Hakala K., Pinta E., Huttunen,S., Pluta P., Gador A.,Lonnberg H. and Skurnik M. Yersiniophage phiR1–37 is a tailed bacteriophage having a 270 kb DNA genome with thymidine replaced by deoxyuridine. Microbiology., 2005 151, 4093–4102.
[4] Yan N., O’Day E., Wheeler L.A., Engelman A. and Lieberman J. HIV DNA is heavily uracilated, which protects it from autointegration. Proc. Natl. Acad. Sci. U.S.A. 2011 108, 9244–9249.
[5] Pettersen H.S., Galashevskaya A., Doseth B., Sousa M.M., Sarno A., Visnes T., Aas P.A., Liabakk N.B., Slupphaug G., Saetrom P. et al. AID expression in B-cell lymphomas causes accumulation of genomic uracil and a distinct AID mutational signature. DNA Repair (Amst), 2015 25, 60–71.
[6] Muha V., Horvath A., Bekesi A., Pukancsik M., Hodoscsek B., Merenyi G., Rona G., Batki J., Kiss I., Jankovics F. et al. Uracil-containing DNA in Drosophila: stability, stage-specific accumulation, and developmental involvement. PLoS Genet. 2012 8,e1002738.
[7] Galashevskaya A, Sarno A, Vågbø CB, Aas PA, Hagen L, Slupphaug G, Krokan HE. A robust, sensitive assay for genomic uracil determination by LC/MS/MS reveals lower levels than previously reported. DNA Repair (Amst). 2013 Sep;12(9):699–706.
[8] Lari SU, Chen CY, Vértessy BG, Morré J, Bennett SE. Quantitative determination of uracil residues in Escherichia coli DNA: Contribution of ung, dug, and dut genes to uracil avoidance. DNA Repair (Amst). 2006 Dec 9;5(12):1407–20.
[9] Horváth A, Vértessy BG. A one-step method for quantitative determination of uracil in DNA by real-time PCR. Nucleic Acids Res. 2010 Nov;38(21):e196.
