|
|
BMe Research Grant |
|
George A. Olah Doctoral School of Chemistry and Chemical Technology
Department of Inorganic and Analytical Chemistry, MTA-BME “Lendület” Chemical Nanosensors Research Group
Supervisor: Dr. Gyurcsányi E. Róbert
Development and Application of Nanopore-based Counters
Introducing the research area
Recently outbroken global epidemics have further enhanced efforts for the development of novel analytical methods in diagnostics. Reliable identification of infections requires fast, accurate, ‘easy to use’ and cost-effective applications. Hopefully, our sensor can overcome this challenge. We develop a nanopore-based particle counter which characterizes synthetic and biological nanoparticles.
Brief introduction of the research place
Although the Chemical Nanosensors Research Group was founded only 3 years ago, a former group had already worked in domestic and international cooperative arrangements on the development of selective methods for the detection of biological markers and chemical components. Amongst other interesting topics, we detect and analyze clinically relevant nanoparticles with nanopore-based sensors.
History and context of the research
The key to inhibiting contagious diseases is fast identification and quarantine, however, false negative tests could have devastating effects. Typically, direct virological methods are based on virus cultivation in cell cultures or on the detection of genetic material via nucleic acid-based amplification. None of these methods are specifically applicable for fast tests in the field. [1]
Bill&Melinda Gates foundation aims at global eradication of poliovirus by vaccinating young children worldwide. Identifying poliovirus is especially challenging, because symptoms are not obvious. The drastic consequence, paralysis is unique, but only occurs with a small part of infections. Selective biosensors detecting the intact virus could solve this problem. My research work connects to this project. We develop a cost-effective method considering viruses as biological nanoparticles.
The sensor adapts the Coulter principle [2], but instead of the micropores applied in cell counters, nanopores of similar size as the target particles are integrated into it. [3] In this method, two electrodes are dipped into two compartments connected by a single pore. Voltage bias connected to the electrodes results ionic current with an intensity depending on electrolyte conductivity and nanopore geometry. [4]
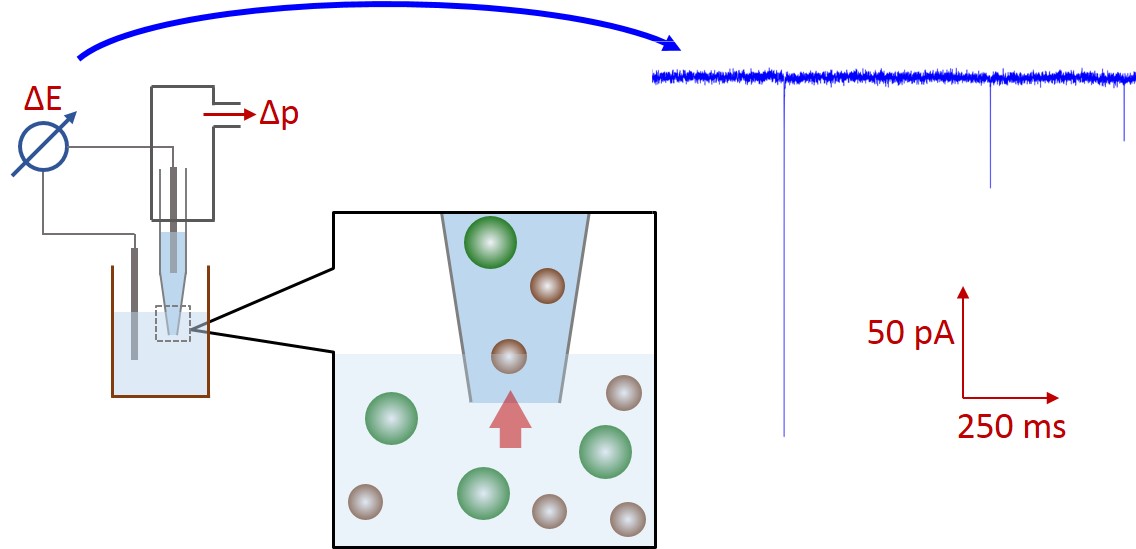
Figure 1: Schematic of the resistive-pulse sensing method.
Particles exclude the volume-equivalent electrolyte amount from the nanopore. Therefore, during particle translocation the pore resistance is increased, so the measured current drops. The amplitude, frequency and width of these current pulses depend on particle size, concentration and velocity. This method detects every single particle, therefore provides stochastic information about the sample. A nanopore is capable of detecting nanoparticles sized 60 % of the aperture or smaller. [5] Bigger particles could clog the hole, while much smaller particle cause current drops get lost in electronic noise.
The research goal, open questions
Adapting the Coulter counter principle requires appropriate sized sensing zone for nanoparticles. We needed a nanopore fabrication method to achieve the polio size range, i.e. 24–30 nm according to literature. The protein capsid of the virus is icosahedral, so it is practically a sphere-like object. [6]
As we had to optimize our sensing method first, we fabricated pores applicable for measuring bigger polystyrene nanoparticles. We subsequently decreased pore diameter to detect smaller ones, too. The measured current amplitudes relate to particle size, while the frequency of translocation events refers to particle concentration.
Commercially available resistive-pulse sensing instruments use concentration and size standards for nanoparticle characterization. As calibration could be challenging in some cases, we investigated a possible way to characterize nanoparticle suspensions in a calibration-free approach.
The main issue with analyzing biological samples (typically serum or environmental sample) is the separation of the target from the sample matrix without significant loss. So we investigated the effect of different sample preparation methods on particle size-distribution and concentration of model polystyrene solutions.
After optimizing our method, we extended our experiments to virus particles derived from vaccines and cell cultures.
Methods
Nanopores were fabricated with Sutter P-2000 micropipet puller. This instrument heats and softens the proper quartz capillary under well-controlled conditions. As the quartz softens enough a programmatic pulling force is applied to produce two conical nanopipets with identical geometries. (Fig 2) Through different parameter settings the diameter of nanopipets is controllable. Compared to other fabrication methods, this is a relatively fast and cost-effective way to analyze particle samples. I only had to change the pulling force to prepare pipets with different diameter.

Figure 2: Schematic of nanopipet pulling
I filled nanopipets with electrolyte solution adjusted to suitable ionic strength and surfactant concentration. We integrate the pipet into the compatible holder, so Ag/AgCl dips into the solution and the pressure can be controlled. In this way, we can control the nanoparticle translocation velocity and clear out any objects from the aperture with an intensive pressure-pulse in case of clogging.
Although there are well-defined models to describe current drops caused by particles translocating cylindrical pores, the state of art lacks simple analytical expressions for RPS in conical pores. Typically, they assume that particle volumes and current amplitudes are proportional and calibrate with known standards, or use some approximations like assuming the end of the tip is cylindrical.
In order to replace particle standards we had to determine the connection between the geometry and resistance of the nanopipets. A numerous set of capillaries were pulled changing the pulling force. In each case one of the two identical pipets were measured with SEM, while the other was filled with electrolyte to characterize its resistance. We determined the R = f (dtip, α, ρ) function, so later we can characterize nanopipet geometry (half-cone angle, diameter) by resistance measurements.
My colleague simulated the current change during the translocation of an insulating sphere through the nanopore. Different particle to pore diameter ratios and half-cone angles were used. Basically, the model defines a polynomial surface, which enables converting the maximal relative current drops into the diameters of the translocating sphere-like nanoparticles using the nanopipet geometry determined from the electrical resistance. [TP1]
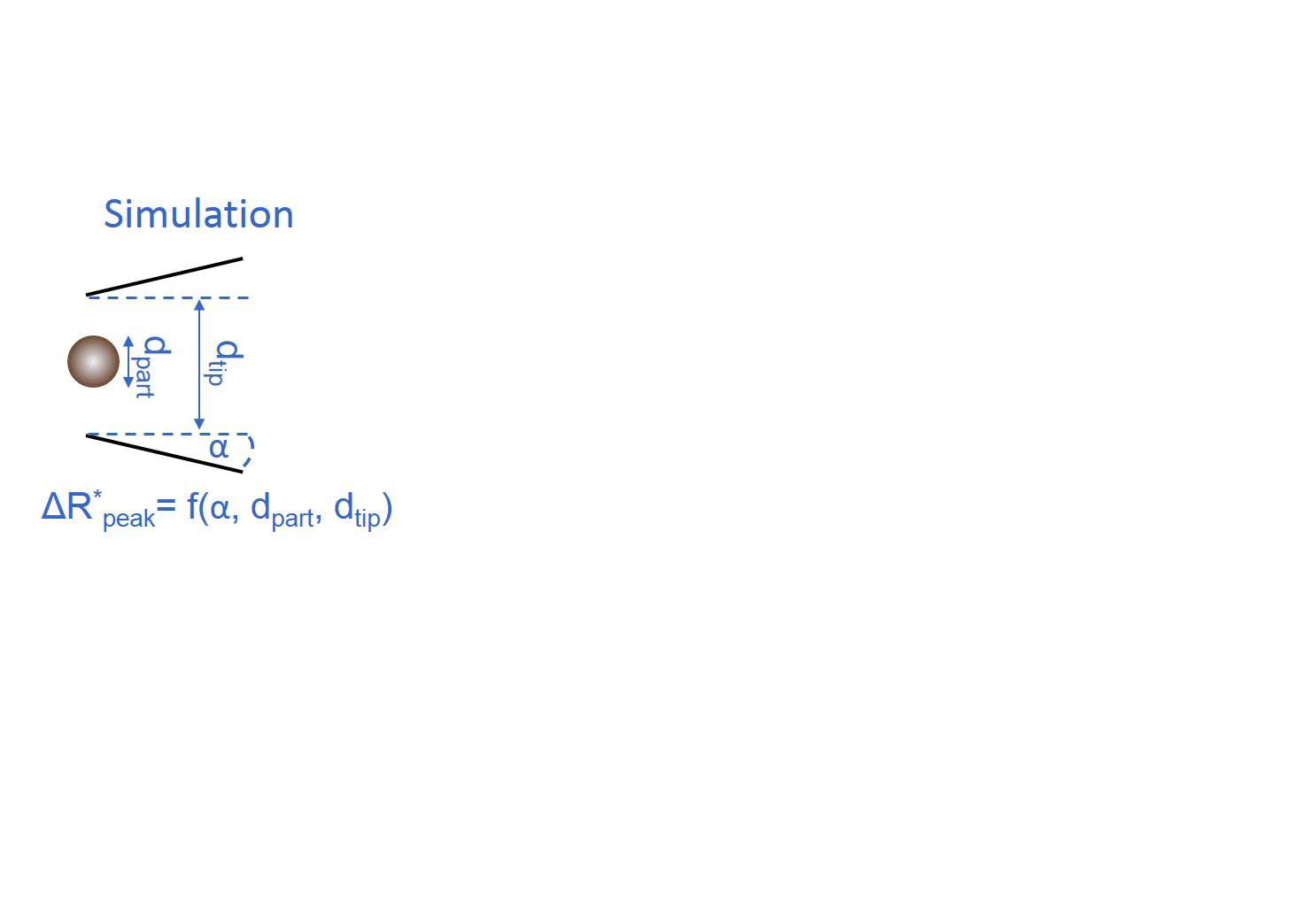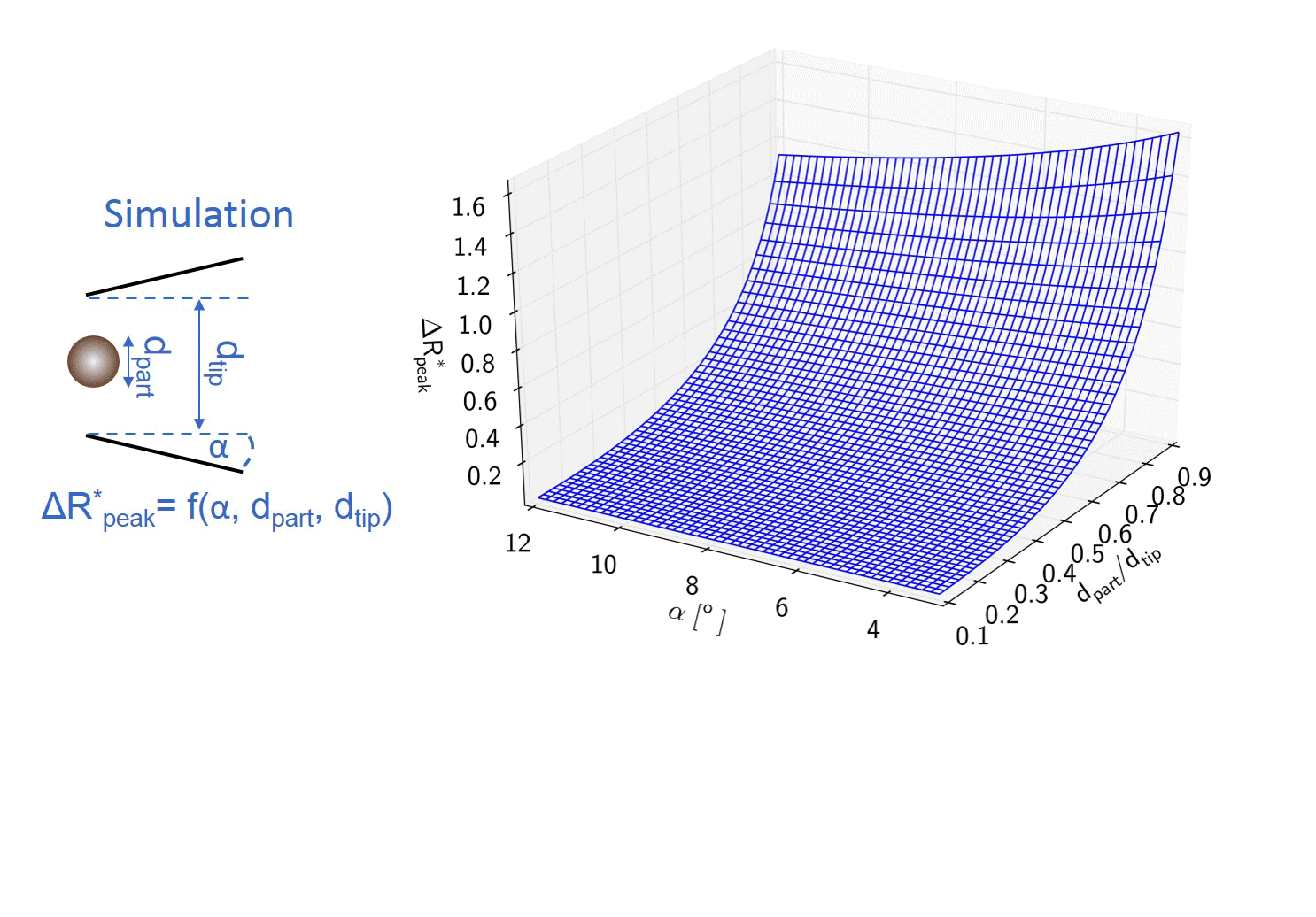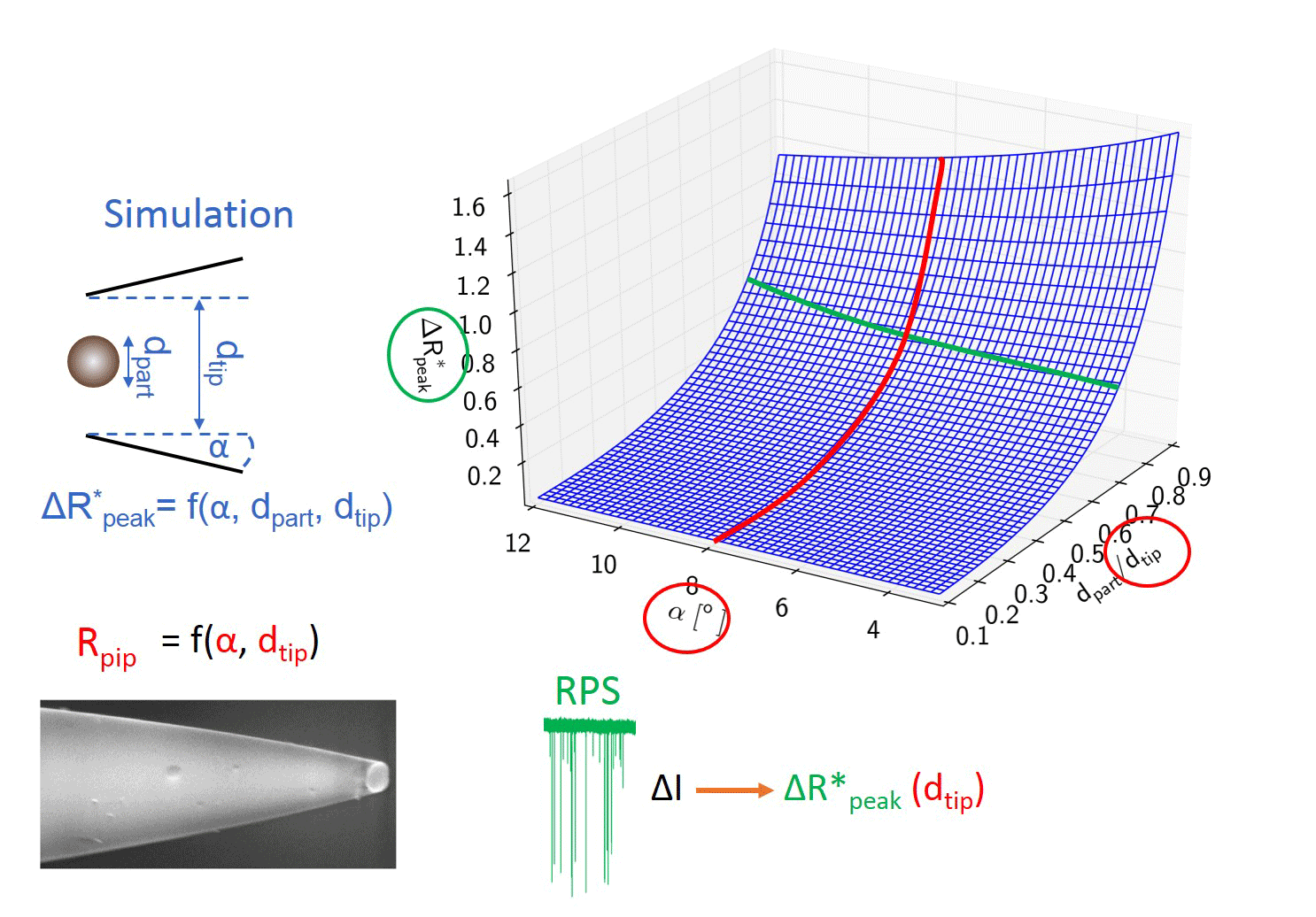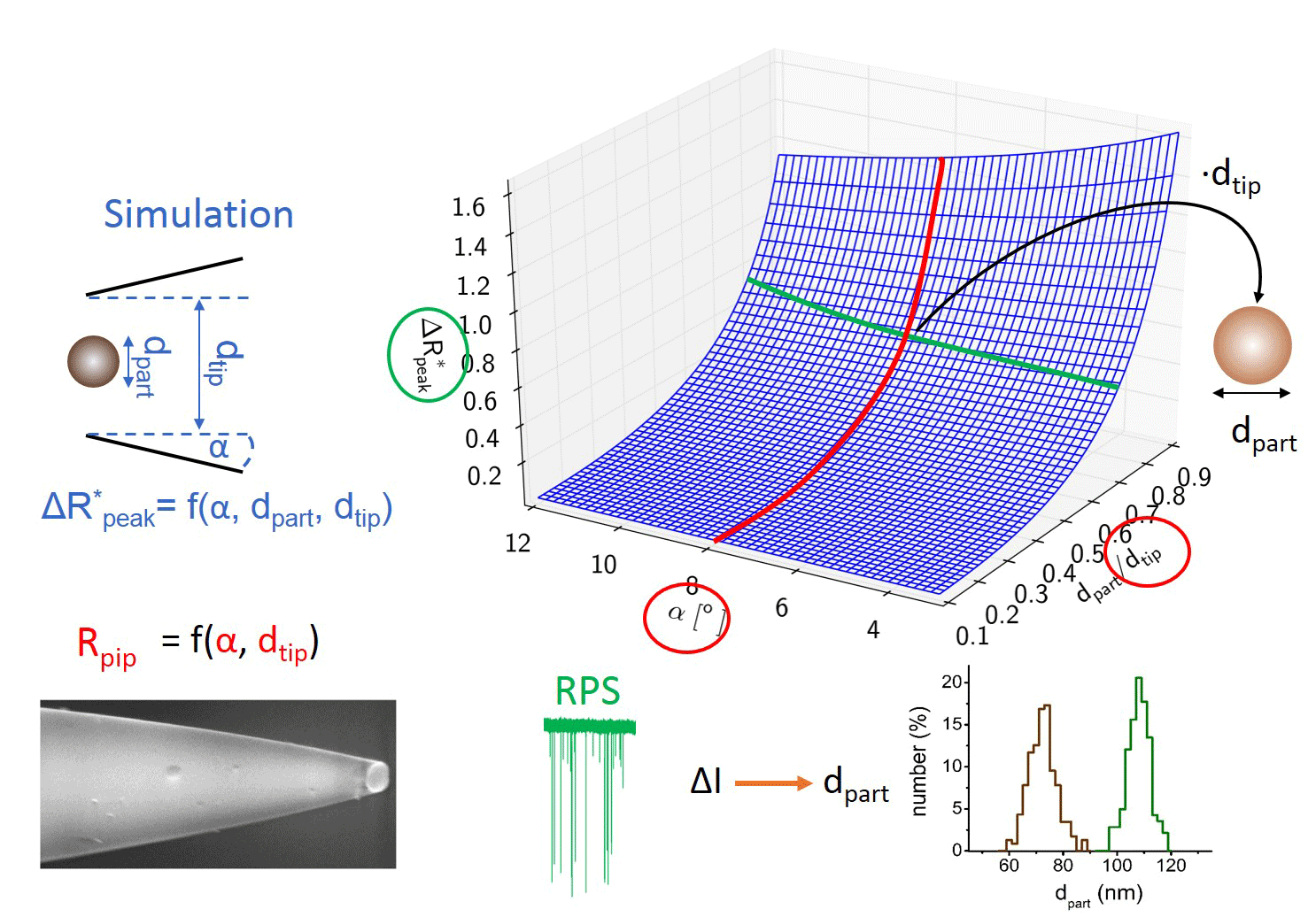
Figure 3: Flip chart of the calibration-less sizing method. [TP1]
I validated the nanoparticle size distributions measured by this calibration-free sizing method via dynamic light scattering(DLS), scanning electron microscopy(SEM), and nanoparticle tracking analysis(NTA). [TP1]
In contrast with commercial Coulter counters the flow rates through nanopores are too small to determine the translocated sample volume. Therefore, event frequency is not convertible to concentration. Typically concentration standards aid quantifying particle samples. To avoid the need for standards, we use the nanopipet geometry to estimate the hydrodynamic resistance of the pore. Via pressure-control in the pipet, we measure event frequencies (f) at different pressures (p). Using the following expression, we determine the nanoparticle concentration from the slope of the frequency - pressure linear function:
![]()
Particle concentrations determined this way were in good agreement with the standard concentrations provided by manufacturers. [TP1]
Results
By changing the pulling force in the instrument’s software I managed to fabricate nanopipets applicable for counting different nanoparticles. During my research I worked out a protocol to produce nanopores for counting particles in the 40–400 nm size range. This enabled the group in a later phase to characterize polioviruses with the nanopipet-based sensor. (Fig 4)
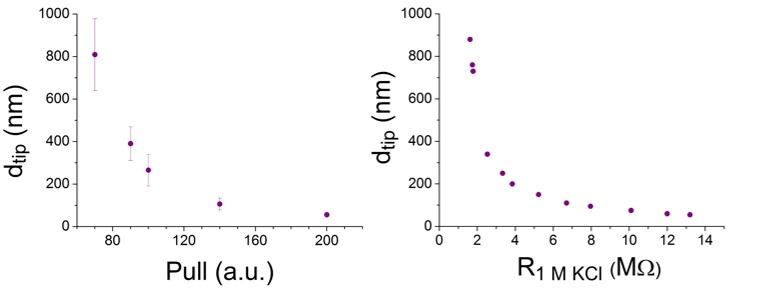
Figure 4: The relationship between nanopore size and pulling force. Nanopipets were measured by SEM and the resistance of the identical pair in 1 molar KCl.
I characterized a large set of nanopipets with electron microscopy or measured their resistance in 1 molar KCl. A nanopipet can only be used either in electron microscopy or in resistive-pulse sensing, so the identical pairs were characterized with different methods. (Fig 4)
The sensing zone broadens for conical geometry broadens, and shortens, too. Therefore, particle translocation typically results in several hundred µs long, triangle-shaped pulse. We evaluate the maximal relative current change and full width at half maximums of these signals. (Fig 5)
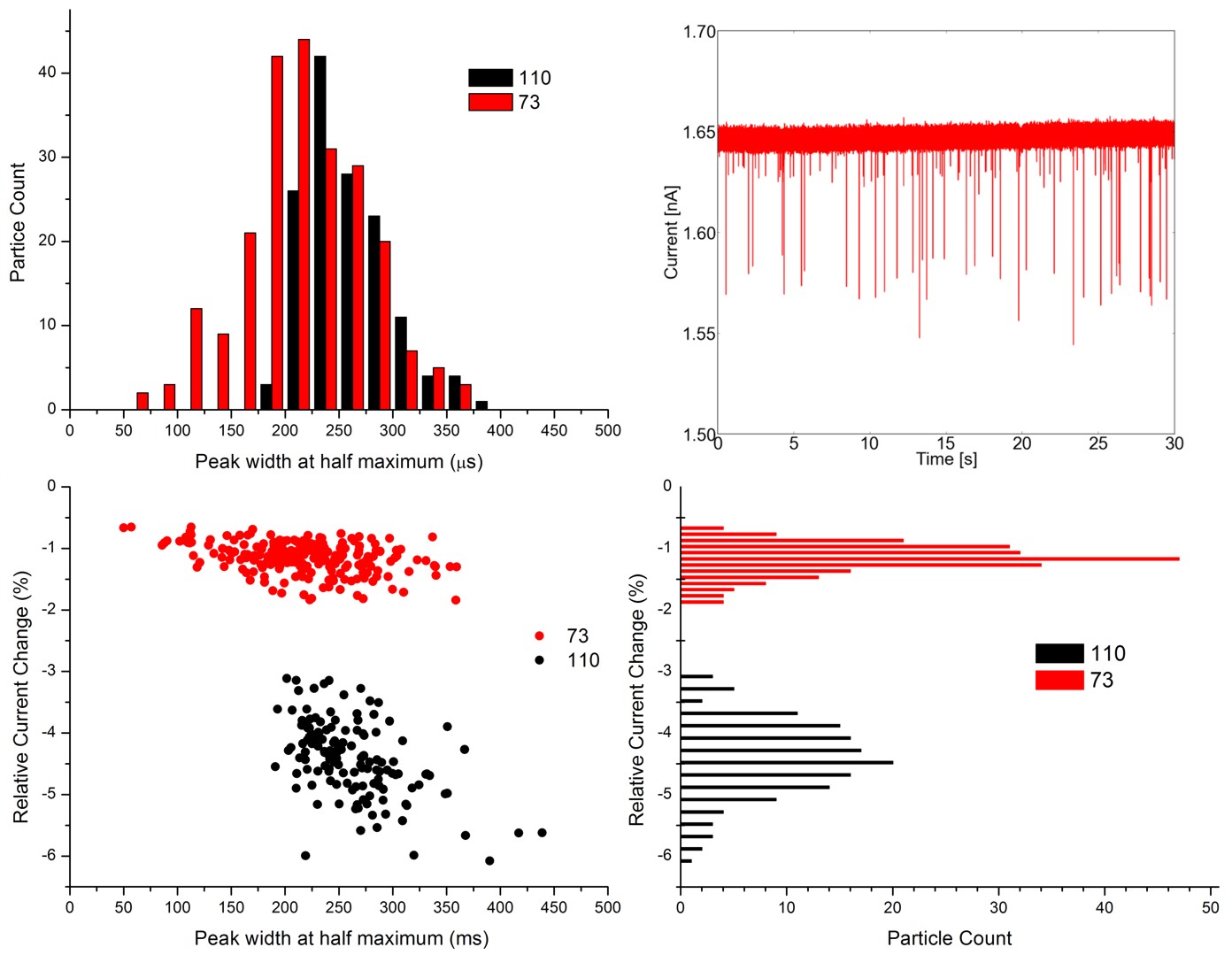
Figure 5: RPS current signals caused by 73 and 110 nm diameter particle mixture, and the evaluated signals on histograms.
By measuring mixtures of different sized monodisperse nanoparticle solutions I managed to prove that resistive-pulse sensing can resolve relatively small size heterogeneities in liquid phase, even where DLS fails. This technique matches electron microscopy in terms of resolving power.
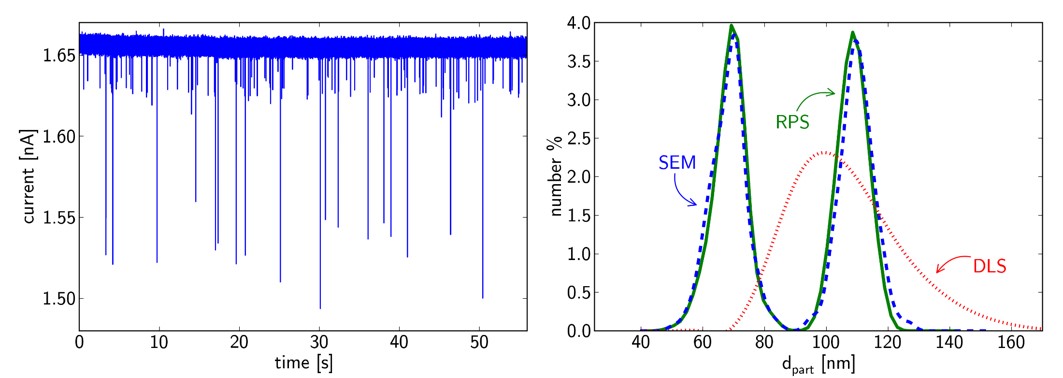
Figure 6: Converted RPS signals of 73 and 110 nm diameter particle mixture, and the same sample evaluated by SEM and DLS. [TP1]
Measuring exact particle concentrations in polydisperse colloid solutions is possible if baseline resolution is reached. The calibration-less quantitation method we proposed can determine the concentration of all resolved particle-distribution. I performed filtration experiments and monitored the concentration and size changes with our calibration-less methods: [TP2]
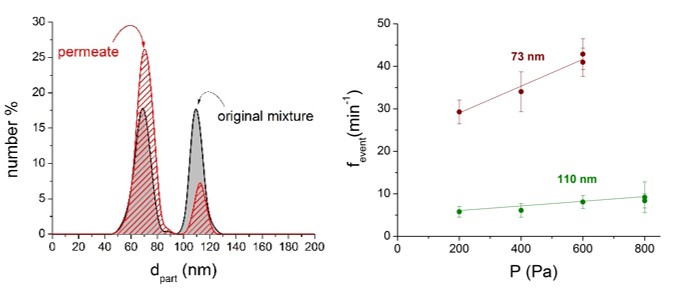
Figure 7: Nanoparticle distribution in a 73 and 110 nm diameter particle mixture before and after membrane filtration. The fevent – P slopes were measured in the permeate. The slopes are proportional to the actual particle concentrations of the sample. [TP2]
At a later stage of my research these studies proved to be useful when I had to characterize rotavirus samples.
Expected impact and further research
The biggest advantage of our proposed methods is that size and concentration standards are not necessary. One drawback of single pore sensing is clogging caused by nonspecific adsorption. Clogged particles in the sensing zone can change baseline current, increase noise and obstruct further particle translocation. Without measuring standards the possibility of these unwanted events decrease significantly.
My further research goal is selectively modulating the signals caused by specific nanoparticles, viruses practically. This way selective counting should enable distinguishing similar sized nanoparticles.
Publications, references, links
[TP1] Terejánszky P, Makra I, Fürjes P, Gyurcsányi RE. Calibration-less sizing and quantitation of polymeric nanoparticles and viruses with quartz nanopipets. ANALYTICAL CHEMISTRY 86:(10) pp. 4688–4697. (2014) IF: 5.636, IC: 9 (doi:10.1021/ac500184z)
[TP2] Terejánszky P, Makra I, Lukács A, Gyurcsányi RE. Nanopipet-Based Resistive Pulse Sensing to Follow Alterations in the Size and Concentration of Nanoparticles During Membrane Filtration. ELECTROANALYSIS 27:(3) pp. 595–601. (2015) IF: 2.471, IC: -
[TP3] Makra I, Terejánszky P, Gyurcsányi RE. A method based on light scattering to estimate the concentration of virus particles without the need for virus particle standards. METHODSX 2: pp. 91–99. (2015) IF: -, IC: 1
References
[1] van den Kieboom CH, van der Beek SL, Mészáros T, Gyurcsányi RE, Ferwerda G, de Jonge MI. Aptasensors for viral diagnostics. TRAC-TRENDS IN ANALYTICAL CHEMISTRY 74: pp. 58–67. (2015)
[2] Coulter WH. Means for counting particles suspended in a fluid. US PATENT No. 2656508 (1953).
[3] Gyurcsányi RE. Chemically-modified nanopores for sensing. TRAC-TRENDS IN ANALYTICAL CHEMISTRY 27:(7) pp. 627–639. (2008)
[4] Luo L, German SR, Lan WJ, Holden DA, Mega TL, White HS. Resistive-Pulse Analysis of Nanoparticles. ANNUAL REVIEW OF ANALYTICAL CHEMISTRY 7:(1) pp. 513–535. (2014)
[5] Bayley H, Martin CR. Resistive-Pulse Sensing - From Microbes to Molecules. CHEMICAL REVIEWS 100:(7) pp. 2575–2594. (2000)
[6] Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM.The Structure of the Poliovirus 135S Cell Entry Intermediate at 10-Angstrom Resolution Reveals the Location of an Externalized Polypeptide That Binds to Membranes. JOURNAL OF VIROLOGY 79:(12) pp. 7745–7755. (2015)
[7] Vogel R, Willmott G, Kozak D, Roberts SG, Anderson W, Groenewegen L, Glossop B, Barnett A, Turner A, Trau M. Quantitative Sizing of Nano/Microparticles with a Tunable Elastomeric Pore Sensor. ANALYTICAL CHEMISTRY 83:(9) pp. 3499–3506. (2011)
[8] Makra I, Gyurcsányi RE. Electrochemical sensing with nanopores: A mini review. ELECTROCHEMISTRY COMMUNICATIONS 43: pp. 55–59. (2014)
Links
